
Haldane kinetic study on biodegradation of phenol -A comprehensive review
Veluru Sridevi 1![]() , Husam Talib Hamzah 1
, Husam Talib Hamzah 1![]()
![]() , Nabil Majd Alawi 2
, Nabil Majd Alawi 2![]() , D. Divya Teja 1
, D. Divya Teja 1![]() , Venkata Rao Poiba 1
, Venkata Rao Poiba 1![]() , Bandi Spandana
1
, Bandi Spandana
1![]() , Husam Salah Mahdi 3
, Husam Salah Mahdi 3![]()
1 Department of Chemical Engineering,
College of Engineering, Andhra University, Visakhapatnam-530003. Andhra
Pradesh. India
2 Department
of Chemical Engineering, University of Technology, Baghdad, Iraq
3 Department of Computer Science and
System Engineering, College of Engineering, Andhra University,
Visakhapatnam-530003. Andhra Pradesh, India
|
|
ABSTRACT |
||
|
The chemical moreover petroleum industries are responsible for the production of a diverse range of organic contaminants that are extremely hazardous. As a result, these industries have contributed to the accumulation of damaging impacts on the surrounding environment. These companies' wastewater typically contains aromatic organic chemicals, which are notoriously difficult to degrade through natural processes and, as a result, are found to be pervasive in the environment. Being the straightforward units for an extensive variety of organic substances, In industries such as oil refining, production of phenol and the various derivatives of it, pharmaceuticals, productions of resins, textile dyes, paints, disinfectants, petrochemicals, and paper mills, phenol and its derivatives are used, and as a result, The effluents produced by these industries often contain phenol as well as derivatives of phenol. The existence of phenolic compounds in water systems is associated with significant increases in the likelihood of adverse health effects being experienced by both human beings and other organisms. In light of this, the elimination of such potentially hazardous substances has garnered a significant amount of focus in recent decades. The removal of phenolic pollutants from aquatic environments by biodegradation is a technique that is both environmentally friendly and economical. For the purpose of optimizing procedure process, building bioreactor systems, and scaling up microbial wastewater treatment procedures to fulfil the requirements of the effluent quality standard, having an understanding of the kinetics of microbial growth and biodegradation is absolutely essential. The current study concentrates on a number of different research publications on Haldane kinetic models, which are utilised to Describe the processes involved in the growth of microbes on phenol. |
|||
|
Received 14 December 2022 Accepted 15 January 2023 Published 31 January 2023 Corresponding Author Husam Talib Hamzah, alwatanaliraqi@gmail.com DOI 10.29121/granthaalayah.v11.i1.2023.4993 Funding: This research
received no specific grant from any funding agency in the public, commercial,
or not-for-profit sectors. Copyright: © 2023 The
Author(s). This work is licensed under a Creative Commons
Attribution 4.0 International License. With the
license CC-BY, authors retain the copyright, allowing anyone to download,
reuse, re-print, modify, distribute, and/or copy their contribution. The work
must be properly attributed to its author.
|
|||
|
Keywords: Biodegradation, Growth Kinetics, Phenol,
Toxicity, Haldane Model |
|||
1. INTRODUCTION
Most of the wastewater that is produced by different types of companies contains phenolic compounds, which frequent type of organic pollutant. In overall, petrochemicals almost (2.8–1220 mg/L), and coke oven plants around (28–1200 mg/L), while coal mining between (9–6800 mg/L) and petroleum oil refineries almost (6–500 mg/L) Which sectors of the economy are primarily at blame for the discharge of phenolic contaminants into the environment that was around them Cetinkaya and Ozdemir (2018), Barik et al. (2021). Likewise, products from industries such as the pharmaceutical, leather, and resin synthesis industries, textile, pulp and paper, paint, moreover industries devoted to the processing of wood hold phenolic compounds variable between 0.1–1600 mg/L Priyadharshini, and Bakthavatsalam (2019).
Phenol is a protoplasmic poison that can frequently be found in industrial wastewater. Its origins can be traced back to coking plants, dyes, varnishes, medications, and insecticides Eyal et al., (2019). Phenol is a common contributor to the failure of wastewater treatment plants, primarily as a result of its high toxicity to living things and general chemical stability Wei et al. (2016), Nawawi et al. (2017). Therefore, The Environmental Protection Agency of the United States has identified phenol being counted among the 126 most significant pollutants Nawawi et al. (2020). The phenol concentration in drinking water shouldn't be higher than 1.0 micrograms per litre at most Anurova et al. (2019), Panigrahy et al. (2020). Every living thing has a fundamental requirement to consume chemicals in order to obtain carbon or energy. Though, it is a widely acknowledged fact that over the course of centuries, cells on earth have adapted to be able to consume these naturally occurring biochemicals, the creation of numerous organic types that are either unaffected to or unable to be mineralized by living organisms, Magharbeh et al. (2021). The elimination of phenolic contaminants through biodegradation is a process that is both environmentally friendly and effective Panigrahy et al. (2022).
On the other hand, due to the eco-friendliness of biological treatments, such as pure or mixed cultures of microorganisms, these kinds of treatments have also been used, bio-mineralization and efficient moreover comprehensive with regard to cost. Phenolic chemicals can be utilised by bacteria, yeast, and fungi. Even though it is poisonous and inhibits the growth of substrates, phenol can assist as a foundation of carbon and energy for a number of dissimilar strains of bacteria that genetically related to classes of Pseudomonas, Aureobasidium, Bacilli, Klebsiella, Ochrobactrum, Rhodococcus, etc, Santos et al. (2019). It is absolutely necessary to have knowledge on the kinetics of the biodegradation of harmful chemicals in order to optimise the removal efficiency and process control. It is a well-established fact that phenolic compounds themselves, have the ability to thwart the degradation of other compounds of phenol, particularly at high level of concentrations. Therefore, the growth kinetics of microorganisms are typically described through the use of the Haldane model. Growth in biomass can be clear as an increase in the quantity of cellular elements as well as the structure of the cell, which is accompanied by an increase in the size of the cell as well as the number of cells. The lag, exponential, stagnant, and death phases are some of the transitions that it goes through Peng et al. (2018). Taking into consideration the vast variety of phenolic component concentrations that are found in industrial wastewaters.
the goal of this review is to Research progression of phenol's growth as well as its biodegradation at cumulative concentrations in order to obtain a knowledge for the impact that inhibition on growth and thus their biodegradation performance. However, to the best of our knowledge, there are no reviews have studied bacterial growth models as well as the kinetics of phenol degradation via Haldane kinetic model.
2. TOXICITY OF
PHENOLIC COMPOUNDS
Phenolic pollutants pose a significant hazard to human health and are known to be carcinogenic. Additionally, they are responsible for a great deal of damage to the ecosystem. In addition, phenolics have a well-deserved reputation for being both genotoxic and endocrine disruptive substances. Compounds with the phenolic structure, like as chlorophenols, nonylphenols, 4-tert-octylphenol, and (BPA) which are public endocrine-disrupting chemicals (EDCs). These chemicals have the potential to inhibit normal hormonal processes, in a variety of organisms, and lead to major hormonal disruption as well as potential health risks (Spataro et al., 2019).
Phenol is toxic for wide variety of aquatic creatures even at inferior concentrations of mg/L, and it causes problems with the smell and taste of the water when it is present Liu et al. (2016), Duan et al. (2018), Noszczynska and Piotrowska-Seget (2018). An excessive amount of phenol exposure consequence in issues for the central nervous system, paralysis, failure of the liver, lack of appetite, rashes, difficulties speaking, digestive disorders, vomiting, morbidity, and a loss of weight are all potential side effects, Cancer is another potential complication. Because of its rapid penetrability and absorption through the skin, as well as through inhalation and eating, it has a lethal effect Liu et al. (2009). It has the potential to cause serious irritation to both the respiratory system and the eyes. The disposal of phenol into the environment is likely to become increasingly difficult as a consequence of increased financial burdens or the production of more hazardous by-products. Before phenol can be discharged into the environment, it is necessary to lower the concentration of phenol down to the desired norms utilising processes that are both physical and chemical in nature as well as biological Al-Asoufi et al. (2017). Though The majority treatments are ineffective to accomplish the point of phenol breakdown, rather than that, transform it into a diverse phase, which results in pollution and toxic by-products. The chemical and physical processes are both included are frequently expensive, and the majority of these treatments unsuccessful to convert phenol into a different phase. However, the biodegradation of phenol is an alternative that is both more beneficial to the environment and more cost-effective. As a direct consequence of this, the application of biological phenol treatment has evolved into an important step in the process of pollution control Khraisheh et al. (2020).
3. BIOLOGICAL
TREATMENT PROCESSES INVOLVING MICROBES
The elimination of phenolic pollutant substances by microbes is one strategy that has the potential to be utilised in the process of eliminating and detoxifying the hazardous pollutants that are existing in polluted areas. Phenolic pollutants like phenol, ethoxylates, nitrophenol, alkylphenol, chlorophenol and cresols, that possible eliminated effectively by a diverse community of microorganisms including fungus, yeasts, microalgae, and bacteria Barik et al. (2021). Microorganisms of a wide diversity have been successfully isolated. from various polluted waterways for the purpose of biodegradation of phenolic pollutants, for instance:
Achromobacter sp.,Rhodococcus sp., Pseudomonas sp., Acinetobacter sp.(SA01), Acinetobacter sp., Bacillus sp., Gulosibacter sp., Arthobacter sp., Halomonas sp., Achromobacter sp., Panigrahy et al. (2020), Barik et al. (2021). Though, few studies have been conducted on the microbiological remediation of large amounts of phenolic pollutants found in actual industrial effluent.
4. MICROORGANISMS IN PHENOL BIODEGRADATION
Xenobiotics that are both toxic and dangerous, which have a composition that is diverse from that of chemicals that are found in nature, and further hard to degrade. This is because they have a different chemical composition. In contrast, in recent years, a wide variety of bacteria that depend on xenobiotics for their continued existence have been discovered Bhatt et al. (2007). Organic matter can be broken down either aerobically or anaerobically, and the method that is used depends on the microbe's capacity to flourish in the given conditions. Lucas et al. (2008), Shah et al. (2008), Lika and Papadakis (2009). Although both aerobic and anaerobic microbes are eligible for breaking down phenol, the aerobic procedures are the ones that are most commonly used Ucun et al. (2010), Melo et al. (2005), Dash et al. (2009). Because they reproduce more quickly and typically convert organic substances to inorganic compounds (CO2, H2O), aerobic bacteria are more effective than anaerobic microbes at decomposing hazardous materials Mrozik et al. (2010). In addition to this, aerobic processes are favoured because of the low costs that are linked with this alternative Ruiz-Ordaz et al. (2001). Because of these factors, there is a relatively low level of attention for usage of anaerobic bacteria for the breakdown of phenols.
5. MECHANISM OF PHENOL BIODEGRADATION
The process of energy conversion that occurs during microbial metabolism is maintained by a number of different reactions; hence, it is a source of ultimate energy. The activity of enzymes, which are uniquely suited to carry out a given kind of chemical reaction, is what guides the metabolic processes. Agarry et al. (2008). Existence of molecular oxygen necessary for process of biodegradation because it kickstarts the attack of the enzymatic nature on the aromatic rings. The process of adding hydroxyl groups to the phenol ring is a typical step in the phenol metabolic pathway, forming catechol by the enzyme phenol hydroxylase, after that, unshroud the ring through ortho- (as well named β-ketoadipate pathway) or meta-oxidation Lika and Papadakis (2009), Jiang et al. (2006) In the metabolic pathway that leads to phenol breakdown, phenol hydroxylase is the first enzyme that is involved.
6. FACTORS AFFECTING BIODEGRADATION OF PHENOLS
There are many different factors that have the potential to influence the ability of microbes to degrade substances or their metabolism by either inhibiting or promoting the growth of the organisms Trigo et al. (2009). The factors include temperature, pH, aeration and agitation, concentration of substrate, and characteristics of pollutants based on physical Nair et al. (2008), El-Naas et al. (2009). To achieve highest possible rate of degradation of the organic chemical of choice, each of these parameters should be fine-tuned for the organism that was chosen. Among the many aspects of phenol biodegradation, one of the most essential is the determining the optimal concentration of the substrate, because it is well accepted that phenol itself can hinder the process of phenol biodegradation carried out by microbial cells, and this inhibition is most noticeable at larger concentrations of the phenol.
7. KINETIC STUDIES
Studies of metabolic kinetics can give a comprehensive examination of rendering and mechanistic features of in situ bioremediation. In order to simulate and make accurate predictions regarding the behavior of microbial degradation, a large number of different kinetics models have been developed. These models have been used in a variety of contexts. There are three distinct stages involved in the process of microbial biodegradation Wen et al. (2020), The first stage consists of a beginning phase during which there is no growth rate, and The next stage is triggered once the growth rate (µ) reaches its maximum acceleration after a certain time, followed by a brief period of delay. In the last phase, there is no longer any decline in the rate of degradation Fan et al. (2004). The highest possible rate of specific biomass growth (µmax) occurs during the process of biodegradation, maximum growth rate (Rm), lag time (λ), saturation constant for substrate (Ks), maximum yield coefficient (Y) are crucial aspects to comprehend in order to get a handle on the biodegradability, microorganisms' affinities for substrates, as well as their compatibility with them.
Kinetics studies of biodegradation reactions provide a measurement of how efficiently the microbial system is functioning. A better understanding of these dynamics will contribute to an improvement in both the control of the procedure, as well as its effectiveness in phenol elimination Agarry et al. (2008). Several kinetic models were utilised in order to adequately represent the process of growth of microbes on phenol and its dynamics. Taking into the rate of growth in biomass, as well as account material balance, and rate of substrate operation (in mg/l.hr) can be characterized by Equation 1 and Equation 2, as follows:
![]()
Equation 1
![]() Equation
2
Equation
2
where Y is the yield of cell mass (g/g) = dX/dS, X represent biomass concentration (mg/l), S represent concentration of substrate (mg/l); kd is the decay coefficient (hr−1); and µ specifific growth rate (hr−1) Tsai and Juang (2006). The Monod model and the Haldane (Andrew's) model are two of the models for biodegradation of phenol that are utilised by researchers the world over.
The understanding kinetics of growth moreover substrate degradation of hazardous compounds is crucial criterion for constructing plants of wastewater treatment. The development kinetics of the microorganism are an essential piece of information that must be possessed before one can have an idea of the possible of the organism to degrade phenol. Keeping in mind the importance of determining the kinetics of growth, For the purpose of describing the dynamics of microbial development, growth inhibitory models are utilised whenever there is a growth-inhibiting substrate present, such as phenol Banerjee and Ghoshal (2010). For the design of biological remediation procedures, the biokinetic parameters that are found from the growth kinetic model that provides the finest fit to the experimental data are required inputs with optimized the operating conditions for the purification of wastewater that has been polluted with phenol Sahoo et al. (2011), Banerjee and Ghoshal (2010). The concentration of biomass that was derived from the batch tests was examined using kinetic models in order to estimate the kinetic parameters Shourian et al. (2009), Cui et al. (2017).
8. DETERMINATION OF GROWTH KINETICS
When applying Haldane's kinetic model, it is necessary to consider phenol to be a growth-inhibiting compound. For the purpose of illustration growth kinetics of inhibitory substrates like phenol, the developed model for the growth kinetics by Haldane was utilised because of its mathematical simplicity and widespread acceptance Wang et al. (2010), Yan et al. (2005). The following are the Equation 3, Equation 4 describes Haldane's inhibitory growth kinetics:
![]()
Equation 3
Where µ-Specific growth rate (day-1), x0- Initial concentration of biomass (gL-1), - x1 -Final concentration of biomass (gL-1), t- reaction time (day).
![]()
Equation 4
Where, µ -Specific growth rate (day-1); x- substrate concentration (gL-1), µmax - maximum specific growth rate (day-1), ks - half saturation constant (gL-1), kί - substrate inhibition constant (gL-1), the kinetic parameters ks, kί, µmax, were determined by nonlinear regression analysis using MATLAB R2016a.
The many additional substrate inhibition models were used in the analysis in order to find out about various kinetic parameters, (Aiba model, Yano & Koga model, Teisser model and Webb model). The following Equation 5, Equation 6, Equation 7, Equation 8 are used in calculations of substrate inhibition kinetic model:
Aiba model:
![]()
Equation 5
Teisser model:
![]()
Equation 6
Yano and Koga model:
![]()
Equation 7
Webb model:
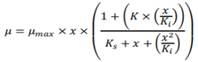 Equation 8
Equation 8
The rate of specific degradation q (day-1) was intended by using results of experiments on phenol degradation, the following equation Equation 9 was developed:
![]()
Equation 9
Where S0 and x are phenol and biomass concentration (g/L) at
reaction time t (day).
9. RECENT RESEARCH ON HALDANE MODEL
Yuzhe et al. (2022) isolated Candida tropicalis sp. exhibiting a significant capacity for biodegradation to phenolic compounds, even in high concentration existence or in an acidic situation. Evaluation of the phenol's biodegradation was carried out using subsequent concentrations around (10–1750 mg/L), The performance of the strain quite fine in terms of its ability to biodegrade. The highest possible specific growth rate 0.660 h−1 and the specific rates of biodegradation 0.47 mg (phenol) [(mg (VSS) h]−1.
Haitham Qaralleh et al. (2022) sought to do research on the phenol's potential for biodegradation by the bacterium that encourages the growth of plants R. nepotum. as the main substrate, there were a total of six distinct phenol concentrations used as the starting point. The capability of cells to biodegrade phenol were found to be considerably impacted by the growing conditions. After incubation for 36 and 96 hours at a pH of 7 and a temperature of 28 degrees Celsius, this bacterium had quickest growth rate and the maximum phenol biodegradation of any tested. biodegradation rate significantly advanced at 700 mg/L, the greatest concentration out of a total of six that was tested. After incubation for a period of fewer than 96 hours, greater than 90 percent of phenol (700 mg/L) removed. When it comes to defining the connection between the beginning level of phenol concentration and growth rate, the Haldane model has proven to be the most reliable. According to the forecast made by the equation of Haldane, the maximum specific growth rate, half-saturation coefficient, and Haldane's growth kinetics inhibition coefficient are 0.7161 h-1, 15.8 (ppm), and 292 (ppm), respectively. The experimental results were effectively modelled using the equation of Haldane, which resulted in a decrease in the SSR (sum of squared errors) to 3.8 x10 3.
Khleifat et al. (2022) recognized plant growth stimulating bacterium C. flaccumfaciens
for kinetics of phenolic compound development
and its subsequent biodegradation and inhibition coefficient (Ki),
half-saturation coefficient (Ks), extreme specific growth rate (µmax) for predicted values for the
phenol-dependent growth kinetics as: 329 (mg/L), 9.14 (mg/L), and 1.05 (h -1 )
respectively, According to what is known as the Haldane model of inhibition,
The growth kinetics proposed by Haldane. Through a sum of squared error (SSR)
of 1.36 X 10 -3, The Haldane equation has excellent compatibility with
the actual data. Enhanced Gombertz model also
successfully predicts the trends of phenol biodegradation. Starting
concentration of phenol had an effect on both the lag
time and rate biodegradation of phenol; the concentration of phenol increased. C. flaccumfaciens growth and
phenol biodegradation was most successful when it was carried out at pH 7 and
temperature of 28degree Celsius incubation.
Magharbeh et al. (2021) studied the degradation of phenol with Bacillus simplex under various circumstances of development. The principal substrate was phenol, and it was employed in each of its six possible starting concentrations. The most optimal growing conditions for this organism, as well as the highest level of phenol biodegradation it can achieve at pH 7, temperature of 280 degrees Celsius, with durations ranging from 36 to 96 hours. The results of the GC-MS screening of the sample taken from the bacterial culture displayed additional breakdown of the catechol was taking place by 1,2-dioxygenase produce a cis, cis-mucconic acid via ortho-pathway and/or by 2,3-dioxygenase into 2-hydroxymucconic semialdehyde via meta-pathway. Maximum rate of biodegradation was apparent at 700 mg/L initial phenol concentration. Around 90 percent of the phenol (700 mg / L) was eliminated in a period of incubation that lasted fewer than 96 hours have passed. It was found to be true that the Haldane model provided the best explanation for the link that existed between the concentration of phenol at the beginning of the experiment and the specific growth rate. The following is the parameters that it is possible to acquire by the use the Haldane equation: 1.05 h−1, 9.14 ppm, and 329 ppm for Haldane’s maximum specific growth rate, the half-saturation coefficient, and the Haldane’s growth kinetics inhibition coefficient, respectively. The experimental data were well described by the Haldane equation, which minimised the sum of squared error (SSR) to 1.36×10−3.
Wen et al. (2020) centred on dynamics of a single, unmixed bacteria strain Rhodococcus sp. SKC, extracted from soil that was polluted with phenol, with the primary purpose of degrading phenol, which serves as its source of carbon and energy in an aqueous medium. Phenol biodegradation kinetics including maximum rate of phenol degradation, lag phase, highest possible growth rate (Rm), and highest possible yield coefficient (Y) for each Si (initial concentration of phenol, mg/L) were adequate with Gompertz and Haldane models of substrate inhibition (R2 > 0.9904, RMSE < 0.00925). These parameters' values when operating under ideal conditions µmax = 0.30 h−1, Ks = 36.40 mg/L, and Ki = 418.79 mg/L, that refers to phenol concentration that causes inhibition was 418.79 mg/L. Through investigation of its relationship to different bacteria types, Rhodococcus sp. SKC displayed a good yield factor and phenol resistance throughout its development.
Priyadharshini and Bakthavatsalam (2019) investigated, microalgae DEE 01 were separated from the municipal waste water treatment facility that dealt with coal gasification, and their growth as well as their phenol use behaviour was studied, and recognized as Chlorella pyrenoidosa based on 18SrRNA sequence analysis (KX686118). The percentage reduction in concentration of phenol across all 4 samples, on average (0.2, 0.4, 0.6 and 0.8 g/L) by C. pyrenoidosa is 97.4% under resident intermediate conditions at pH-8. Additionally, it may be capable of degrading phenol in the effluent coal gasification. In order to match the experimental data, the Haldane inhibitory growth kinetics model was utilised, and the kinetic parameters that were found were as follows: Extreme specific growth rate- µmax = 0.7123 day−1 , half saturation constant- Ks=0.1899 g/L ;tolerance to toxicity -Ki=0.6898 g/L with R 2=0.9916. The kinetic study of C. pyrenoidosa is experiential to possess supreme specific growth and degradation rate, (Ki) and high phenol affinity (Ks).
Arutchelvan and Atun (2019) discovered a strain of bacterium that is able using phenol as the only foundation of carbon, was obtained from the different samples of soil, gathering from the area surrounding an industrial treatment facility for wastewater that services a phenol manufacturing unit. According to the results of biochemical analyses and 16S rRNA created on the results of the sequencing, the organism was known as Serratia marcescens. Highly poisonous phenolic chemical can be degraded by the organism by an optimal concentration of 2500 mg/L in 120 h and pH 8. Additionally, the organism was effective and had a broad pH and temperature tolerance in addition to a very strict lag phase when dealing with greater concentrations influent. Among two diverse models combined to Explain how organism's growth rate is determined, the Haldane’s model fits (R2=0.930) to a high degree of contentment with kinetic constants falling somewhere in the range of µmax = 0.05 - 0.095 h-1; Ks = 8.49 - 16.1 mg/L; Kj - 1154.75 - 1700.68 mg/L.
Peng et al. (2018) described Haldane model for the rate of development of both PNP-acclimated activated sludge and phenol. The estimate of Haldane's kinetic parameters are as follows: maximum specific growth rate, µmax, constant of saturation, Ks, and inhibition constant, Ki, were discovered. The lesser ratio of Ks/Ki for degradation of phenol compared to that of PNP biodegradation shows the increased resistance of the activated sludge that was acclimated with phenol to the inhibition caused by its substrate. The kinetics of phenol degradation could be described using the pseudo zeroth-order equation, regardless of the initial compound concentration. Though, the equation that was supposed to describe the degradation of PNP was inadequate, and this was attributable to the fact that PNP is more hazardous than phenol.
Bera et al. (2017) isolated Stenotrophomonas acidaminiphila, Brevibacterium sp. and Brucella sp. from a sample of sludge taken at one of the refineries in Assam, which is located in India. It was discovered that the mixed culture had the ability to break down the highest possible concentration of phenol 1000 mg L-1 for 96 h whereas highest possible specific growth rate (µmax) detected at 100 mg/L. Optimum values for pH 6.5 and temperature 37 Celsius to achieve complete phenol degradation. Phenol is broken down by the mixed culture through ortho-cleavage pathway by the creation of midway (cis, cis-muconate) It was identified by the use of spectrophotometry at 260 nm. The validity of the experimental results was established by comparing the growth and substrate usage curves to the simulated dynamic profiles that corresponded to those curves, gotten by solving equation of Haldane via MATLAB R2015a with µmax = 0.155 h-1 and KI = 400 mg L-1.
10. CONCLUSION
The low substrate level presents challenges for the traditional physicochemical treatment of industrial wastewaters including chemicals such as phenol, increasing use of chemicals, development of potentially harmful by products, and an increase in the overall expense of the operation. It would appear that biological treatment is the way to go in order to handle these types of industrial wastewaters. The Haldane substrate inhibition models were utilised in order to provide an explanation for the substrate inhibition caused by phenol as well as the growth kinetics of the culture. The biokinetics constants were analysed using models, and the results showed that the culture had a good tolerance for the conditions and was growing well, moreover whole substrate degradation.
Future work
Future work should be focused on isolating and identifying novel microorganisms as a priority and evaluating the phenol-degrading capabilities of the samples, both laboratory and field conditions, with a focus on large-scale pilot experiments and field-scale applications being taken into special care.
CONFLICT OF INTERESTS
The authors certify that the publishing of this paper does not involve any potential conflicts of interest on their part.
ACKNOWLEDGMENTS
This work was supported by Department of Chemical Engineering, Andhra University, Visakhapatnam. Andhra Pradesh, India.
REFERENCES
Agarry, S. E., Durojaiye, A. O., Yusuf, R. O., Aremu, M. O., Solomon, B. O., and Mojeed, O. (2008). Biodegradation of Phenol in Refinery Wastewater by Pure Cultures of Pseudomonas Aeruginosa NCIB 950 and Pseudomonas Fluorescence NCIB 3756. Int. J. Environ. Pollut., 32, 3-11. https://doi.org/10.1504/IJEP.2008.016894.
Agarry, S. E., Durojaiye, A. O., and Solomon, B. O. (2008). Microbial Degradation of Phenols: A Review. Int. J. Environ. Pollut., 32, 12-28. https://doi.org/10.1504/IJEP.2008.016895.
Al-Asoufi, A., Khlaifat, A., Tarawneh, A.A., Alsharafa, K., Al-Limoun, M., and Khleifat, K. (2017). Bacterial Quality of Urinary Tract Infections in Diabetic and Non-Diabetics of the Population of Ma'an Province, Jordan. Pakistan J. Biol. Sci., 20(4), 179-188. https://doi.org/10.3923/pjbs.2017.179.188.
Anurova, M. N., Bakhrushina, E.O., Demina, N.B., and Panteleeva, E.S. (2019). Modern Preservatives of Microbiological Stability. Pharm. Chem. J., 53, 564-571. https://doi.org/10.1007/s11094-019-02038-4.
Arutchelvan, V., and Atun, R. C. (2019). Degradation of Phenol, an Innovative Biological Approach. Adv Biotech & Micro., 12(2), 555835. https://doi.org/10.19080/AIBM.2019.12.555835.
Banerjee, A., and Ghoshal, A.K. (2010). Phenol Degradation by Bacillus Cereus: Pathway and Kinetic Modeling. Bioresour. Technol., 101, 5501-5507. https://doi.org/10.1016/j.biortech.2010.02.018.
Barik M., Das C.P., Verma A.K., Sahoo S., Sahoo N.K. (2021). Metabolic Profiling of Phenol Biodegradation by an Indigenous Rhodococcuspyridinivorans Strain PDB9T N-1 Isolated from Paper Pulp Wastewater. Int. Biodeterior. Biodegrad., 158, 105168. https://doi.org/10.1016/j.ibiod.2020.105168.
Bera, S. Roy, A. S., and Mohanty, K. (2017). Biodegradation of Phenol by a Native Mixed Bacterial Culture Isolated from Crude Oil Contaminated Site. International Biodeterioration & Biodegradation, 121, 107-113. https://doi.org/10.1016/j.ibiod.2017.04.002.
Bhatt, P., Kumar, M. S., Mudliar, S., and Chakrabarti, T. (2007). Biodegradation of Chlorinated Compounds-A Review. Crit. Rev. Env. Sci. Technol., 37, 165-198. https://doi.org/10.1080/10643380600776130.
Cetinkaya and Ozdemir (2018). Cetinkaya A.Y., Ozdemir
O.K. Phenol Removal from Synthetic Solution Using Low Pressure Membranes Coated
with Graphene Oxide and Carbon. Chem. Pap., 72, 327-335. https://doi.org/10.1007/s11696-017-0282-9.
Cui, P., Mai, Z., Yang, S., and Qian, Y., (2017). Integrated Treatment Processes for Coalgasification Wastewater with High Concentration of Phenol and Ammonia. J.Cleaner Production , 142, 2218-2226. https://doi.org/10.1016/j.jclepro.2016.11.056.
Dash, R. R., Gaur, A., and Balomajumder, C. (2009). Cyanide in Industrial Wastewaters and its Removal: A Review On Biotreatment. J. Hazard. Mater., 163, 1-11. https://doi.org/10.1016/j.jhazmat.2008.06.051.
Duan, W., Meng, F., Cui, H., Lin, Y., Wang, G., Wu, J. (2018). Ecotoxicity of Phenol and Cresols to Aquatic Organisms: A Review. Ecotoxicol. Environ. Saf., 157, 441-456. https://doi.org/10.1016/j.ecoenv.2018.03.089.
El-Naas, M. H., Al-Muhtaseb, S., and Makhlouf, S. (2009). Biodegradation of Phenol by Pseudomonas Putida Immobilized in Polyvinyl Alcohol (PVA) gel. J. Hazard. Mater., 164, 720-725. https://doi.org/10.1016/j.jhazmat.2008.08.059.
Fan, Y., Wang, Y., and Qian, P.Y. (2004). Optimization of Phthalic Acid Batch Biodegradation and the Use of Modified Richards Model for Modelling Degradation. Int. Biodeterior. Biodegrad., 53, 57-63. https://doi.org/10.1016/j.ibiod.2003.10.001.
Jiang, H. L., Tay, S. T., Maszenan, A. M., and Tay, J. H. (2006). Physiological Traits of Bacterial Strains Isolated from Phenol-Degrading Aerobic Granules. FEMS Microbiol. Ecol., 57, 182-191. https://doi.org/10.1111/j.1574-6941.2006.00114.x.
Khleifat, K., Magharbeh, M., Alqaraleh, M., Al-Sarayrah, M., Alfarrayeh, I., Al Qaisi, Y., Alsarayreh, A., & Al-kafaween, M. A. (2022). Biodegradation modeling of phenol using Curtobacterium flaccumfaciens as plant-growth-promoting bacteria. Heliyon, 8(9), 10490. https://doi.org/10.1016/j.heliyon.2022.e10490.
Khraisheh, M., Al-Ghouti, M.A., and AlMomani, F.P. (2020). Putida as Biosorbent for the Remediation of Cobalt and Phenol from Industrial Waste Wastewaters. Environ. Technol. Innovat., 20, 101148. https://doi.org/10.1016/j.eti.2020.101148.
Kurzbaum, E., Raizner, Y., Kuc, M. E., Kulikov, A., Hakimi, B., Kruh, L. I., & Menashe, O. (2020). Phenol Biodegradation by Bacterial Cultures Encapsulated in 3D Microfiltration-Membrane Capsules. Environmental Technology, 41(22), 2875–2883. https://doi.org/10.1080/09593330.2019.1587005.
Lika, K., and Papadakis, I. A. (2009). Modeling the Biodegradation of Phenolic Compounds by Microalgae. J. Sea Res. 62, 135-146. https://doi.org/10.1016/j.seares.2009.02.005.
Liu, Q. S., Liu, Y., Show, K.Y., Tay, J.H. (2009). Toxicity Effect of Phenol on Aerobic Granules, Environ. Technol., 30(1), 69-74. https://doi.org/10.1080/09593330802536339.
Liu, Z., Xie, W., Li, D., Peng, Y., Li, Z., Liu, S. (2016). Biodegradation of Phenol by Bacteria Strain Acinetobacter Calcoaceticus PA Isolated from Phenolic Wastewater. Int. J. Environ. Res. Publ. Health, 13(3), 300. https://doi.org/10.3390/ijerph13030300.
Lucas, N., Bienaime, C., Belloy, C., Queneudec, M., Silvestre, F., and NavaSaucedo, J. (2008). Polymer Biodegradation: Mechanisms and Estimation Techniques. Chemosphere, 73, 429-442. https://doi.org/10.1016/j.chemosphere.2008.06.064.
Magharbeh, M.K., Khleifat, K.M., Al-kafaween, M.A., Saraireh, R., Alqaraleh, M., Qaralleh, H., Al-Tarawneh, A., Al-limoun, M.O., El-Hasan, T., Hujran, T., Ajbour, S.H., Jarrah, N., Amonov, M., Al-Jamal, H.A.N. (2021). Biodegradation of Phenol by Bacillus Simplex: Characterization and Kinetics Study. Applied Environmental Biotechnology, 6(2), 1-12. https://doi.org/10.26789/AEB.2021.02.001.
Melo, J. S., Kholi, S., Patwardhan, A. W., and D'Souza, S. F. (2005).
Effect of Oxygen Transfer Limitations in Phenol Biodegradation. Process
Biochem., 40, 625-628.
https://doi.org/10.1016/j.procbio.2004.01.049.
Mohammad Nawawi, N., Shukor, M. Y., and Ibrahim, A. L. (2018). Characterization of Phenol-Degrading Bacteria: A Review. Selangor Science & Technology Review (SeSTeR), 1(1), 71–83.
Mrozik, A., Cycon, M., and Piotrowska-Seget, Z. (2010). Changes of FAME ' Profiles as a Marker of Phenol Degradation in Different Soils Inoculated with Pseudomonas sp. CF600. Int. Biodeterior. Biodegrad., 64, 86-96. https://doi.org/10.1016/j.ibiod.2009.11.002.
Nair, C. I., Jayachandran, K., and Shashidhar, S. (2008). Biodegradation of Phenol. African Journal of Biotechnology, 7(25), 4951-4958.
Nawawi, T., Fomina, M., and Dumanskaya, T. (2020). A New Rhodococcus Aetherivorans Strain Isolated from Lubricant-Contaminated Soil as a Prospective Phenol-Biodegrading Agent. Appl. Microbiol. Biotechnol., 104, 1-15. https://doi.org/10.1007/s00253-020-10385-6.
Noszczynska, M., and Piotrowska-Seget, Z.
(2018). Bisphenols: Application, Occurrence, Safety, and Biodegradation
Mediated by Bacterial Communities in Wastewater Treatment Plants and Rivers.
Chemosphere, 201, 214-223. https://doi.org/10.1016/j.chemosphere.2018.02.179.
Panigrahy N., Barik M., Sahoo R.K., Sahoo N.K. (2020). Metabolic Profile Analysis and Kinetics Of P-Cresol Biodegradation by an Indigenous Pseudomonas Citronellolis NS1 Isolated from Coke Oven Wastewater. Int. Biodeter. Biodegr., 147, 104837. https://doi.org/10.1016/j.ibiod.2019.104837.
Panigrahy, N., Priyadarshini, A., Sahoo, M. M., Verma, A. K., Daverey, A., & Sahoo, N. K. (2022). A Comprehensive Review on Eco-Toxicity and Biodegradation of Phenolics: Recent Progress and Future Outlook. Environmental Technology and Innovation, 27, 102423. https://doi.org/10.1016/j.eti.2022.102423.
Peng, S. S., Ling, N. S., and Rohana, A. (2018). Kinetics of Biodegradation of Phenol and P-Nitrophenol by Acclimated Activated Sludge. Journal of Physical Science, 29(1), 107-113. https://doi.org/10.21315/jps2018.29.s1.14.
Priyadharshini S.D., and Bakthavatsalam A.K. (2019). A Comparative Study on Growth and Degradation Behavior of C. Pyrenoidosa on Synthetic Phenol and Phenolic Wastewater of a Coal Gasification Plant. J. Environ. Chem. Eng., 7, 103079. https://doi.org/10.1016/j.jece.2019.103079.
Qaralleh, H., Khleifat, K. M., Maha, N., Hajleh, A., Muhamad, O., Al-Limoun, Alshawawreh, Magharbeh , R., M. K., Al-Qaisi, T. S. Farah, H. S., Tayel El Hasan , Al-Tarawneh, A., Aljbour, S. H. (2022). Plant Growth-Promoting Rhizobium Nepotum Phenol Utilization: Characterization and Kinetics. Moath Alqaraleh Journal of Hunan University (Natural Sciences, 49 (4). https://doi.org/10.55463/issn.1674-2974.49.4.11.
Ruiz-Ordaz, N., Ruiz-Lagunez, J. C., Castañon-González, J. H., Hernández-Manzano, E., Cristiani-Urbina, E., & Galíndez-Mayer, J. (2001). Phenol Biodegradation Using a Repeated Batch Culture of Candida Tropicalis in a Multistage Bubble Column. Revista latinoamericana de microbiologia, 43(1), 19–25.
Sahoo, N.K., Ghosh, P.K., and Pakshirajan, K. (2011). Kinetics of 4-bromophenol Degradation Using Calcium Alginate Immobilized Arthrobacter Chlorophenolicus A6. Int J Earth Sci Eng., 4, 663-668.
Santos, V.L., Monteiro, A.S., Braga, D.T., and Santoro, M.M. (2009). Phenol Degradation by Aureobasidium Pullulans FE13 Isolated from Industrial Effluents. J of Hazard Mat., 61(2-3), 1413-1420. https://doi.org/10.1016/j.jhazmat.2008.04.112.
Shah, A. A., Hasan, F., Hameed, A., and Ahmed, S. (2008). Biological Degradation of Plastics: a Comprehensive Review. Biotechnol. Adv., 26, 246-265. https://doi.org/10.1016/j.biotechadv.2007.12.005.
Shourian, M., Noghabi, K.A., Zahiri, H.S., Bagheri, T.,
Karballaei, G., Mollaei, M., Rad, I., Ahadi, S., Raheb, J., Abbasi, H., (2009).
Efficient Phenol Degradation by a Newly Characterized, Pseudomonas Sp. SA01
Isolated from Pharmaceutical Wastewaters. Desalination, 246, 577-594. https://doi.org/10.1016/j.desal.2008.07.015.
Spataro, F., Ademollo, N., Pescatore, T., Rauseo, J., and Patrolecco, L. (2019). Antibiotic Residues and Endocrine Disrupting Compounds in Municipal Wastewater Treatment Plants in Rome, Italy. Microchem. J., 148, 634-642. https://doi.org/10.1016/j.microc.2019.05.053.
Trigo, A., Valencia, A., and Cases, I. (2009). Systemic
Approaches to Biodegradation. FEMS Microbiol. Rev., 33, 98-108. https://doi.org/10.1111/j.1574-6976.2008.00143.x.
Tsai, S. Y., and Juang, R. S. (2006). Biodegradation of Phenol and Sodium Salicylate Mixtures by Suspended Pseudomonas Putida CCRC 14365. J. Hazard. Mater., B138, 125-132. https://doi.org/10.1016/j.jhazmat.2006.05.044.
Ucun, H., Yildiz, E., and Nuhoglu, A. (2010). Phenol Biodegradation in a Batch Jet Loop Bioreactor (JLB): Kinetics Study and Ph Variation. Bioresour. Technol., 101, 2965-2971. https://doi.org/10.1016/j.biortech.2009.12.005.
Wang, L., Min, M., Li, Y., Chen, P., Chen, Y., Liu, Y., Wang, Y., Ruan, R. (2010). Cultivation of Green Algae Chlorella Sp. In Different Wastewaters from Municipal Wastewater Treatment Plant. Applied Biochemistry and Biotechnology, 162, 1174-1186. https://doi.org/10.1007/s12010-009-8866-7.
Wei, X., Gilevska, T., Wetzig, F., and Dorer, C. (2016). Characterization of Phenol and Cresol Biodegradation by Compound-Specific Stableisotope Analysis. Environ. Pollut., 210, 166-173. https://doi.org/10.1016/j.envpol.2015.11.005.
Wen, Y., Li, C., Song, X. and Yang, Y. (2020). Biodegradation
of Phenol by Rhodococcus Sp. Strain SKC: Characterization and Kinetics Study.
Molecules, 25, 3665.
https://doi.org/10.3390/molecules25163665.
Yan, J., Jianping, W., Hongmei, L., Suliang, Y., Zongding, H., (2005). The Biodegradation of Phenol at High Initial Concentration by the Yeast Candida Tropicalis. Biochem. Eng. J., 24, 243-247. https://doi.org/10.1016/j.bej.2005.02.016.
Yuzhe, He., Wang, Z., Li, T., Peng, X., Tang,
Y., and Jia, X. (2022). Biodegradation of Phenol by Candida Tropicalis
Sp.: Kinetics, Identification of Putative Genes and Reconstruction of Catabolic
Pathways by Genomic and Transcriptomic Characteristics. Chemosphere., 308(3),
136443. https://doi.org/10.1016/j.chemosphere.2022.136443.
 This work is licensed under a: Creative Commons Attribution 4.0 International License
This work is licensed under a: Creative Commons Attribution 4.0 International License
© Granthaalayah 2014-2023. All Rights Reserved.