
VALIDATION OF MORPHOLOGICAL CRITERIA FOR CLONAL IDENTIFICATION IN BUDWOOD GARDENS OF RUBBER TREE (Hevea brasiliensis Muell. Arg.) CLONES USING MICROSATELLITE MARKERS
Hervé Michel
Kouakou 1,2,3 ![]()
![]() , Angeline. A. E Elabo 1, Inza Jésus Fofana 2, N’da Désiré Pokou 3, Assanvo Simon-Pierre N’guetta 4, Yedoh Michel Gnagne 1, Arnaud Freddy Yapi 5, Saraka Didier Martial Yao 2, Nafan Diarrassouba 2,
Samuel Obouayeba 6
, Angeline. A. E Elabo 1, Inza Jésus Fofana 2, N’da Désiré Pokou 3, Assanvo Simon-Pierre N’guetta 4, Yedoh Michel Gnagne 1, Arnaud Freddy Yapi 5, Saraka Didier Martial Yao 2, Nafan Diarrassouba 2,
Samuel Obouayeba 6
1 Doctor, Ex-engineer, Genetic Improvement Operation, National
Center for Agronomic Research (CNRA), Research Station of Bimbresso, 01 P.O.
Box 1536 Abidjan 01, Ivory Coast
2 Doctor, Faculty of Biological Sciences,
Department of Biochemistry-Genetics, Pedagogical and Research Unit of Genetics,
University Péléforo Gon Coulibaly (UPGC), P.O. Box 1328 Korhogo, Ivory Coast
3 Doctor, Central Laboratory of Biotechnology,
National Center for Agronomic Research (CNRA), 01 P.O. Box 1740 Abidjan 01,
Ivory Coast
4 Professor, Laboratory of Genetics,
University Félix Houphouët Boigny, 22 P.O. Box 582, Abidjan 22, Ivory Coast
5 Doctor,
Faculty of Biological Sciences, Department of Biology and Plant Physiology,
University Péléforo Gon Coulibaly (UPGC), P.O. Box 1328 Korhogo, Ivory Coast
6 Doctor, Agronomy Physiology Operation, National Center
for Agronomic Research (CNRA), Research Station of Bimbresso, 01 P.O. Box 1536
Abidjan 01, Ivory Coast
|
|
ABSTRACT |
||
|
The inability of almost all rubber tree nursery
growers to distinguish between the different rubber tree clones in a budwood
garden (JBG) often leads to clonal mixtures. This makes the application of
latex harvesting technology unsuitable, which negatively impacts the yield
and / or the economic life of the plantations. To overcome this problem, a
clonal identification method based on the use of seven (7) morphological
criteria in JBG (budwood garden) was developed. Knowing that the
morphological criteria are influenced by the environment, the visual
recognition of the plants seems approximate or inefficient. In order to check the effectiveness of the morphological
criteria, five (5) rubber tree genotypes from two JBGs (budwood gardens) were
characterized both morphologically and by microsatellite markers.
Characterization by microsatellites was carried out on twelve (12) loci and
involved a sample of 25 individuals per genotype. This first study on the use
of microsatellite markers revealed that three (3) microsatellite markers
(Hb36, Hb43 and Hb110) best discriminate the genotypes (GT1, PB217, IRCA41,
IRCA230 and IRCA331) cultivated in Côte d'Ivoire. It also showed that an
individual declared non-compliant by the morphological method has 89% chance
of being genetically non-compliant, one could be mistaken by 11% in declaring
an individual non-compliant while it is genetically compliant (type I error).
In addition, an individual declared compliant by the morphological method has
a 76% chance of being genetically non-compliant, one could also be mistaken
by 24% in declaring compliant while it is genetically non-compliant (type II
error). |
|||
|
Received 19 September 2022 Accepted 20 October 2022 Published 04 November 2022 Corresponding Author Hervé
Michel Kouakou, mikydollard53@gmail.com DOI10.29121/granthaalayah.v10.i10.2022.4826 Funding: This research
received no specific grant from any funding agency in the public, commercial,
or not-for-profit sectors. Copyright: © 2022 The
Author(s). This work is licensed under a Creative Commons
Attribution 4.0 International License. With the
license CC-BY, authors retain the copyright, allowing anyone to download,
reuse, re-print, modify, distribute, and/or copy their contribution. The work
must be properly attributed to its author.
|
|||
|
Keywords: Hevea Brasiliensis, Rubber Tree Clones,
Budwood Garden, Microsatellite Marker, Morphological Criteria, Reliability,
Error |
|||
1. INTRODUCTION
Rubber trees are cultivated around the world mainly from clones grafted onto rootstocks. The latter come from the sowing of the seeds collected in the plantations. The graft is taken from selected plant material and kept in budwood gardens (JBGs). The establishment of a budwood garden (JBG) and the production of planting material requires the use of compliant rubber tree genotypes and professional quality maintenance Compagnon (1986).
Notwithstanding this requirement, the strong demand for planting material has led the rubber sector to liberalize the production of this material. We are witnessing the increasing proliferation of JBGs (budwood gardens) held by individuals who are not trained in their management. The inability of almost all nursery growers and farmers to distinguish between the different rubber tree clones in JBG (budwood garden) and in plantation runs the risk of clonal mixtures. However, clonal mixtures are a major constraint to achieving yield objectives because efficient latex harvesting technologies cannot be applied.
In order to overcome this problem, a study to develop a clonal identification key based on the use of morphological criteria was conducted Gnagne et al. (2014a). This work made it possible to determine in JBG (budwood garden, seven (7) clone identification criteria including three (03) main and four (04) secondary ones Gnagne et al. (2014a). This identification method was based on a multivariate analysis and made it possible to obtain levels of phenotypic variations greater than 80%, with a risk of error of less than 5% within the population of clones recommended in Côte d’Ivoire Gnagne et al. (2014a). However, morphological criteria are often influenced by the environment. The morphological identification key in budwood garden needs to be confirmed by a genetic fingerprinting approach. This study aims at determining the level of reliability of morphological identification criteria in JBG (budwood garden) using microsatellite markers, with a view to their promotion in the sector.
2. REVIEW OF METHODOLOGICAL APPROACHES
2.1. REVIEW OF MATERIAL AND STUDY LOCATION
Table 1 shows the plant material consisted of five (05) clones of Hevea brasiliensis cultivated in Côte d'Ivoire. These included clones GT1, PB 217, IRCA41, IRCA230 and IRCA331. These genotypes were chosen because they are the most cultivated in Côte d'Ivoire.
Table 1
|
Table 1 Characteristics of Hevea Brasiliensis Clones Cultivated in Côte d'Ivoire |
|||||
|
Clones |
Geographical origin and year of
creation |
Genetic origin |
Dry rubber yield (kg/ha) |
Number of stimulations |
|
|
GT1 |
Indonesia (Java) 1930 |
Primary clone (Control in all
large-scale trials) |
2000-2500 |
5-10 |
|
|
PB217 |
Malaysia 1955 |
PB5/51 x PB6/9 |
2500-3000 |
3-8 |
|
|
IRCA41 |
Côte d’Ivoire 1974 |
GT1 x PB5/51 |
2500-3000 |
8-13 |
|
|
IRCA230 |
Côte d’Ivoire 1976 |
GT1 x PB5/51 |
2000-2500 |
|
|
|
IRCA331 |
Côte d’Ivoire 1978 |
GT1 x RRIM600 |
2500-3000 |
|
|
The study was carried out in two budwood gardens located in the towns of Songon (site A) and Dabou (site B) in southern Ivory Coast. The geographical coordinates of site A are (5°00 N; 3°00 W) and those of site Bare (5°19 N; 4°34 W). The climate encountered is of the humid subtropical type with four seasons. It is characterized by a bimodal rainfall regime with two rainy seasons (from April to July and from October to November), and two dry seasons (from December to February or even March and from August to September). The average annual rainfall varies from 1700 to 1800 mm per year. The average monthly temperature is between 25.5 and 27°C Keli et al. (1992). The relief of these areas is dominated by plains whose altitudes vary between 0 and 100 m, sometimes isolating hills of 200 to 300 M Obouayeba (2005). The derived soil cover consists of deep, loose, and well-drained ferrallitic soils (Ferralsols), poor in exchangeable bases and acid reacting (6 ≤ pH ≤ 7.5) Assiri et al. (2015).
2.2. REVIEW OF ASSESSMENT OF THE RELIABILITY OF THE METHOD OF MORPHOLOGICAL IDENTIFICATION
2.2.1. CHARACTERIZATION OF RUBBER TREE GENOTYPES IN BUDWOOD GARDENS USING THE MORPHOLOGICAL METHOD
The plants chosen in the JBGs (budwood gardens) of sites A and B for the observations were those which showed three (03) or four (04) leaf stages with especially the last leaf stage fully blooming. Observations of morphological characters were made on the penultimate leaf stage, i.e., the second or third leaf stage of the plant. The measurements were preferably carried out on the 3rd leaf starting from the bottom upwards. Seven (7) morphological criteria defined by Gnagne et al. (2014a) were used to make this morphological characterization. These included the length of the central petiolule, the color of the leaflets, the arrangement of the leaflets, the shape of the central leaflet, the angle between the petiole and the stem, the presence of nectar and the shape of the axillary bud. The use of these characters made it possible to determine the number of non-compliant plants, those which do not belong to the same group of identified clones.
2.2.2. MOLECULAR CHARACTERIZATION
·
Leaf Sampling
The leaves of healthy individuals were cut and placed in coded envelopes. A first random draw of three individuals was carried out among the non-compliant individuals, then a second one with the remaining compliant individuals. A sample of 25 individuals per clone and per site was kept, the individuals were numbered from 1 to 125 for site A and from 126 to 237 for site B.
·
DNA Extraction
Genomic DNA extraction was performed from 150 mg of fresh leaves per individual. The leaves were initially crushed in liquid nitrogen at -196°C and then incubated in a buffer from the “ZR Plant/Seed DNAMiniPrepTM” extraction kit (ZYMO RESEARCH, USA). The DNA was isolated by successive centrifugations in various solutions then purified on a filtration column according to the protocol established by the kit supplier. The isolated DNA was diluted in 50 µl of pure water and stored at -30°C.
·
DNA Quantification
The extracted DNAs were quantified using a UV Vis 2000 nanodrop (Fisher Thermoscientific, USA) from a volume of 1 µl per sample of crude DNA extract. This volume was used to measure the optical density (OD) at 260 nm (the DNA concentration in ng/µl), 280 nm (the amount of protein contained in the DNA suspension), and the ratio (OD260/OD280) of the extracts. This ratio between these two measurements of OD: OD260/OD280 is a way to assess the quality or purity of our DNA extract. For a ratio around 2, the DNA extract is qualified as better or pure and its use in several amplification techniques is likely to give good results.
·
PCR Amplification of Microsatellites
Twelve (12) pairs of SSR primers (Hb31, Hb32, Hb33, Hb36, Hb43, Hb45, Hb53, Hb55, Hb64, Hb68, Hb78, Hb110) were used to amplify the DNA fragments by PCR using a GeneAmp System 9700 thermal cycler (Applied Biosystem, USA). These SSR markers were chosen from the GenBank database, available at https://www.ncbi.nlm.nih.gov. The amplification was carried out in a 10 μl reaction medium consisting of Mix Dream TaqTM Green PCR Master buffer (1X)'', Taq DNA polymerase (1 Unit), dNTP 0.4 mM each, 4 mM of MgCl2, 0.1 μM of each primer and 10 ng of DNA extract. The PCR was carried out using the primers labeled with fluorochromes IRD700 and IRD800 which are used for visualization on a screen after laser reading.
·
Migration and visualization of amplification
products
The Li-COR 4300 Genetic Analyzer was used for both 6.5% Long Ranger Type Polymer Gel Electrophoresis and amplification product development. The sizes (in number of base pairs) of the alleles of the microsatellite markers visualized were determined by referring to the bands of a size marker (IRD700 and IRD800).
2.2.3. ASSESSMENT OF THE RELIABILITY OF THE FIRST METHOD OF MORPHOLOGICAL IDENTIFICATION
·
Definition of Events
The events were defined according to Giovani (2005), from the declarations of the morphological and molecular method applied to the samples of rubber tree clones, as follows:
1) NCm: “the morphological method declares an individual non-compliant or off-type” event,
2) Cm: “the morphological method declares an individual compliant” event,
3) NCM: “the molecular method declares an individual non-compliant” event,
4) CM: “the molecular method declares an individual compliant” event.
·
Probability of Events or a priori
Probabilities
The probabilities of the events were calculated according to the method of Giovani (2005), following the formula (i):
p= Number of clones of the event/ Total number of clones (1)
·
Use of Conditional probabilities and
Application of Bayes' Theorem
The Bayes conditional probabilities (issued in 1763, modernized in 1958) applied were determined according to the method of Giovani (2005), along two axes:
Axis 1: declaring an individual non-compliant by the morphological method. In this case, the power of the morphological method to declare an individual non-compliant while it is actually declared non-compliant by the molecular method is expressed through probability (ii). The error of declaring an individual non-compliant while it is actually declared compliant by the molecular method (type I error) (iii).
P(NCM/NCm) = p(NCM)
x p(NCm/NCM) / p(NCm) With
p(NCm) ≠ 0 (2)
P(CM/NCm) = 1- P(NCM/NCm) (3)
Axis 2: Declaring an individual compliant by the morphological method. In this case, the power of the morphological method to declare an individual compliant while it is really compliant by the molecular method is expressed through probability (iv). The error of declaring an individual compliant while it is really non-compliant by the molecular method (type II error) (v).
P(CM/Cm) = p(CM) x p(Cm / CM) / p(Cm) With p(Cm)
≠ 0 (4)
P(NCM/Cm)
=1-P(CM/Cm) (5)
The power of the morphological identification test is given through P(NCM/NCm) et P(CM/Cm).
·
Statistical analyses
The individuals detected as non-compliant by the morphological method were used to estimate impurity levels (Ti).
Ti ![]() X 100
X 100
GENETIX software version 4.03, F-STAT version 2.9.3 and Genalex version 6.5 Peakall and Smouse (2012), Darwin version 6.5 Perrier and Jacquemoud-Collet (2006) and STRUCTURE version 2.3.2 Pritchard et al. (2000), were used to process the information contained in the generated genetic data matrix. This software allowed to see the polymorphism of the microsatellites, the allelic richness, intra- and inter-population variability, as well as the genetic structure of the population. The images of the gels stored on the sequencer were analyzed using Xn View v2.2, Jelly v0.1 (Rami, unpublished) and Excel v2019. The STRUCTURE software probabilistically assigns clone samples to genetically distinct groups (K) and estimated proportions for each clone sample in a population.
3. RESULTS
3.1. IMPURITY LEVEL OF ALL JBG (BUDWOOD GARDEN) STRAINS
Impurity level of JBG (budwood garden) strains from
site A
The results show that no non-compliant individual was identified in the plots of clones GT1, IRCA230 and IRCA331. Three (03) strains were identified as non-compliant in the plots of clone IRCA41 as well as in those of clone PB217. The impurity levels of clones IRCA41 and PB217 were 1% and 1.4%, respectively. These impurity levels were less than 5%. The average impurity level on site A was 0.5%, less than 5% shown in Table 2.
Table 2
|
Table 2 Impurity Level of Rubber Tree Clones Determined by the Morphological Identification Method in the Budwood Garden of Site A |
||||||
|
Clones |
GT 1 |
PB 217 |
IRCA 41 |
IRCA 230 |
IRCA 331 |
Avg |
|
Number
of strains described |
290 |
216 |
300 |
309 |
303 |
|
|
Number
of compliant strains |
290 |
213 |
297 |
309 |
303 |
|
|
Number of non-compliant strains |
0 |
3 |
3 |
0 |
0 |
|
|
Impurity
level (%) |
0 |
1.4 |
1 |
0 |
0 |
0.5 |
Impurity
level of JBG (budwood garden) Strains from Site B
No non-compliant individual was identified in the plots of clone IRCA331. An Impurity level of 4% lower than 5% was detected in the plots of clone IRCA331. While in the plots of clones IRCA41, PB217 and IRCA230 impurity levels greater than 5% were detected, respectively 7.1%, 10.7% and 15%. The average impurity level on site B was 7.36%, greater than 5% shown in Table 3.
Table 3
|
Table 3 Impurity Level of Rubber Tree Clones Determined by the Morphological Identification Method in the Budwood Garden of Site B |
||||||
|
Clone |
GT1 |
PB217 |
IRCA41 |
IRCA230 |
IRCA331 |
Avg |
|
Number
of strains described |
222 |
186 |
197 |
186 |
12 |
|
|
Number of compliant strains |
213 |
166 |
183 |
158 |
12 |
|
|
Number of non-compliant strains |
9 |
20 |
14 |
28 |
0 |
|
|
Impurity levels (%) |
4 |
10.7 |
7.1 |
15 |
0 |
7.36 |
3.2. POLYMORPHISM OF MICROSATELLITE MARKERS
Only 6 (six) pairs of primers amplified out of the twelve (12) tested in this study. Among these amplified microsatellite markers, three, or 50%, proved to be polymorphic and discriminating. These included markers Hb43, Hb36 and Hb110 shown in Table 4. A polymorphism with a total of 8 different alleles was identified in the five (05) populations of rubber tree clones using the three (03) microsatellites
Table 4
|
Table 4 Selected Microsatellite Markers, Their Pattern, Their Sequence and Their Hybridization Temperature |
||||||
|
Locus |
GT1 |
PB217 |
IRCA41 |
IRCA230 |
IRCA331 |
Total
different alleles |
|
HB43 |
1 |
2 |
2 |
2 |
1 |
2 |
|
HB36 |
2 |
1 |
1 |
2 |
1 |
3 |
|
HB110 |
1 |
2 |
1 |
1 |
1 |
3 |
|
Total |
4 |
5 |
4 |
5 |
3 |
8 |
|
% |
50 |
62.5 |
50 |
62.5 |
37.5 |
|
|
Average
number of alleles per clone and per locus |
1.33 |
1.66 |
1.33 |
1.66 |
1 |
2.66 |
The number of alleles per locus varied from two (02) for the Hb43 locus to three (03) for the Hb36 and Hb110 loci with an average of 2.66 alleles per locus shown in Table 5.
Table 5
|
Table 5 Number of alleles per locus and per clone |
|||
|
Locus |
Sequence (5’- 3’) |
Size of alleles Observed (pb) |
Temp (°C) |
|
HB 43 |
F: TTGTCTCCCCTTAATTCTGCTCTT |
232 / 238 |
58 |
|
|
R: GTGATCTGCCCATAACTACTCCAT |
|
|
|
HB 36 |
F: AGTGGCCAAGAAAGAATAAAA |
240 / 253 / 255 |
58 |
|
|
R: TACTACCCATCCACCAACCTAA |
|
|
|
HB 110 |
F: ATGCAGCGATGTAGATAAAAGA |
287 / 295 / 301 |
58 |
|
|
R: TCAAGATGTAAGCACCAGAACT |
|
|
The three (3) loci Hb36, Hb43 and Hb110 were used in combination to obtain the typical profiles of the 5 clones grown in Côte d'Ivoire shown in Table 6.
Table 6
|
Table 6 Genotypes of the five (5) rubber tree clones cultivated in Côte d'Ivoire, obtained by combining loci Hb43, Hb36 and Hb110 |
|||
|
Clones |
HB43 |
HB36 |
Hb110 |
|
GT1 |
232 / 232 |
240 / 253 |
295 / 295 |
|
PB217 |
232 / 238 |
240 / 240 |
287 / 295 |
|
IRCA41 |
232 / 238 |
240 / 240 |
295 / 295 |
|
IRCA230 |
232 / 238 |
240 / 253 |
295 / 295 |
|
IRCA331 |
232 / 232 |
240 / 240 |
295 295 |
3.3. ANALYSIS OF THE ELECTROPHORETIC PROFILES GENERATED BY THE THREE POLYMORPHIC AND DISCRIMINATING MARKERS (HB36, HB43, HB110) OF THE 5 RUBBER TREE GENOTYPES
· Electrophoretic profile of clone GT1. This clone was homozygous for markers Hb110 and Hb43 with a 295 Bp band and a 232 Bp band, respectively. GT1 was heterozygous for marker Hb36 with 240 and 253 Bp bands. Eight (8) samples were detected as non-compliant among the 50 strains sampled on the two sites. All non-compliant individuals were identified in Site B JBG (budwood garden) shown on Figure 1.
· Electrophoretic Profile of Clone PB217. Clone PB217 was homozygous for marker Hb36 with a 240 Bp band and heterozygous for markers Hb110 and Hb43 with the observation of two (295 Bp, 287 Bp) and (238 Bp, 232 Bp) bands, respectively. Sample No. 98 (23rd sample) of this clone showed a profile that could not be visualized at the level of marker Hb43. It was therefore not taken into account in the continuation of this study. Among the 49 samples with readable profiles, 12, namely two (02) for site A and ten (10) for site B showed profiles different from the standard profiles defined. They were therefore declared non-compliant shown on Figure 2.
· Electrophoretic Profile of Clone IRCA41. IRCA41 was represented by homozygous samples for markers Hb110 and Hb36 with a 295 Bp band and a 240 Bp band, respectively. They were also heterozygous for marker Hb43 with 238 and 232 Bp bands. No band could be visualized in sample 120 (20th individual) of this clone, at the level of marker Hb43. It was therefore not taken into account in the continuation of this study. Ten (10) samples including four (4) for site A and six (6) for site B were detected non-compliant among 49 individuals with readable profiles shown on Figure 3.
· Electrophoretic Profile of Clone IRCA230. This clone was homozygous for marker Hb110 with a 295 Bp band and heterozygous for markers Hb36 and Hb43 with the observation of two (253 Bp, 2240 Bp) and (238 Bp, 232 Bp) bands, respectively. Sample No. 30 (5th sample) of this clone showed a profile that could not be visualized at the level of marker Hb43, so it was not taken into account in the continuation of this study. Among the 49 readable profiles, five (5) corresponding to samples from site B were declared non-compliant shown on Figure 4.
· Electrophoretic Profile of Clone IRCA331. IRCA331 was homozygous for all Hb110, Hb36 and Hb43 markers with a 295 Bp band and a 240 Bp and 232 Bp band, respectively. Four (4) samples from site B were detected non-compliant among the 37 individuals sampled in the plots of IRCA331 from both sites, shown on Figure 5.
Figure 1
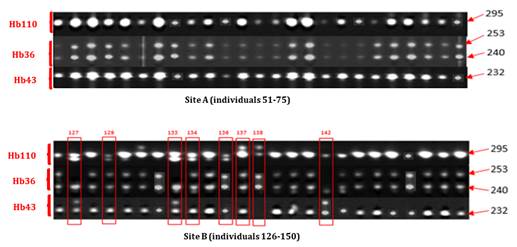
|
Figure 1 Electrophoretic Profiles of 25 Strains of Clone GT1 of Sites A And B Generated By Microsatellites Hb343, Hb36 and Hb110 |
Figure 2
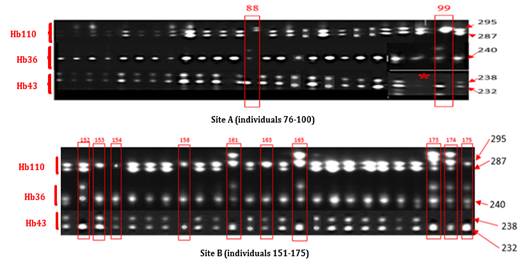
|
Figure 2 Electrophoretic Profiles of 25 Strains of Clone PB217 of Sites A And B Generated By Microsatellites Hb343, Hb36 and Hb110 |
*: No band
Figure 3

|
Figure 3 Electrophoretic Profiles of 25 Strains of Clone IRCA41 Of Sites A And B Generated By Microsatellites Hb343, Hb36 and Hb110 |
*: No band
Figure 4
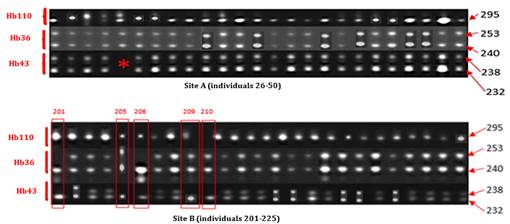
|
Figure 4 Electrophoretic profiles of 25 strains of clone IRCA230 of sites A and B generated by microsatellites Hb343, Hb36 and Hb110 |
*: No band
Figure 5
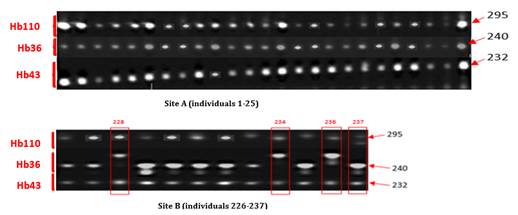
|
Figure 5 Electrophoretic Profiles of 25 Strains of Clone IRCA331 From Site A And 12 Strains From Site B generated by microsatellites Hb343, Hb36 and Hb110 |
3.4. DETERMINATION OF THE POWER OF THE MORPHOLOGICAL IDENTIFICATION TEST
Molecular analysis detected 195 compliant individuals, out of the remaining 234 individuals, that is, 83% of individuals, and 39 non-compliant individuals, that is, 17%. In contrast, the morphological characterization had identified 208 compliant individuals, that is, 89% of individuals and 26 non-compliant, that is, 11% shown in Table 7.
Table 7
|
Table 7 Theoretical number of individuals obtained by comparison of the morphological method with the molecular method and event probabilities |
|||
|
Status |
Number |
Compliant morphological method (Cm) |
Non-compliant morphological method (NCm) |
|
Compliant molecular method (CM) |
195 (83 %) |
188 (96 %) |
7 (4 %) |
|
Non-compliant molecular method (NCM) |
39 (17 %) |
20 (51 %) |
19 (49 %) |
|
Total |
234 |
208 (89 %) |
26 (11 %) |
Among the 208 individuals declared compliant by the morphological method, 188 or 96% were confirmed compliant by the molecular method. The remaining 20 individuals detected compliant by the morphological method were detected non-compliant by the molecular method. Nineteen individuals, with a percentage of 49%, were confirmed non-compliant by the molecular method, out of the 26 individuals detected non-compliant by the morphological method, while the remaining 7 individuals or 4% detected non-compliant by the morphological method were detected compliant by the molecular method, shown in Table 7.
The results from the power of the test revealed that an individual declared compliant by the morphological method has a 76% chance of being compliant by the molecular method. In contrast, an individual declared compliant by the morphological method has a 24% chance of being non-compliant (type II). An individual declared non-compliant by the morphological method has an 89% chance of being non-compliant by the molecular method and an individual declared non-compliant by the morphological method has an 11% chance of being declared compliant by the molecular method shown in Table 8.
Table 8
|
Table 8 Conditional Probabilities and Power of The Morphological Identification Test in JBG (Budwood Garden), Determined Using Bayes' Theorem |
|||
|
Status |
Power of the test (%) |
Error (%) |
Error type |
|
Compliant molecular method (CM) |
76 % |
24 % |
Type II |
|
Non-compliant molecular method
(NCM) |
89 % |
11 % |
Type I |
4. DISCUSSION
The morphological characterization carried out using 7 morphological criteria allowed the detection of non-compliant individuals and, indirectly, the overall impurity level of the plots. This level of impurities was on average 0.5% for site A and 7.36% for site B. Indeed, plots with levels of impurities below 5% showed few non-compliant individuals and those with impurity levels above 5% had more non-compliant individuals. According to Démange et al. (1990), the purity of a budwood garden is measured through the number of compliant individuals likely to be found in these plots. When the impurity level is greater than 5%, this JBG (budwood garden) is considered impure and must be automatically destroyed. The fairly low impurity level obtained on site A indicates that this JBG (budwood garden) is well maintained and usable for an identification study Démange et al. (1990), Gnagne et al. (2014a). Moreover, the molecular profiles of these individuals would make it possible to better purify the JBGs (budwood gardens) of site B which showed a high number of non-compliant plants.
Only markers Hb36, Hb43, Hb110 generated polymorphic profiles out of the twelve (12) markers used to screen our clones. They can therefore be recommended for the molecular characterization of rubber tree clones cultivated in Côte d'Ivoire. However, the validation of the morphological criteria by microsatellite markers indicates that the JBG (budwood garden) morphological identification method appeared to be effective with a power of 76% in declaring an individual compliant, confirmed by the molecular method and 89% in declaring an individual non-compliant, confirmed by the molecular method. It also showed that it is possible to be wrong by 24% in declaring an individual compliant while it is non-compliant according to the molecular method (type II error) and by 11% in declaring an individual non-compliant while it is compliant according to the molecular method (type I error). Indeed, the error made by the morphological method by declaring an individual non-compliant while it is compliant is less serious. Of course, it mistakenly eliminates plants or genotypes useful to breeders for conservation. Moreover, it also determines a degree of confidence in the morphological identification method Kouakou (2017). In contrast, type II error made in a clone by declaring an individual compliant while it is non-compliant is more serious, since it allows a straightforward deviation from the objectives set. In the process of JBG (budwood garden) purification, all non-compliant individuals can be eliminated or regrafted. Under these conditions, it is preferable to declare a genetically compliant individual off-type and eliminate or regraft it rather than declaring a genetically non-compliant individual as compliant, preserving it and even multiplying it by marketing it under a false name Gnagne et al. (2014a). The results obtained certainly show the usefulness of the morphological method, without however excluding that these listed identification errors could be due to the person or the team that carried out the visual inspection. According to Gnagne et al. (2014a), less careful visual inspection in budwood gardens could give much poorer or even mediocre results. This validation indicates that visual inspection is a good method for characterizing and purifying budwood gardens, at least for a small number of well-known rubber tree genotypes with a well-trained technician who regularly practices.
5. CONCLUSION
This study aimed at validating the morphological criteria for clonal identification of five clones in budwood garden using microsatellite markers. It made it possible to determine on plants with 3-4 leaf stages impurity levels on both sites, whose molecular profiles were confirmed by polymorphic markers Hb36, Hb43 and Hb110. The study showed that an individual declared non-compliant by the morphological method has an 89% chance of being non-compliant and an individual declared compliant by the morphological method has a 76% chance genetically compliant. It also showed the limits of the morphological criteria, in particular 11% declaring an individual non-compliant while it is compliant (type I error) and 24% declaring compliant while it is non-compliant (type II error). These errors are relevant clues which demonstrate that morphological characterization seems to be a good method for characterizing and purifying budwood gardens in Ivory Coast. Thus, to better control the purity of JBGs (budwood gardens), an improvement of the morphological criteria is recommended, using a large number of morphological characters.
CONFLICT OF INTERESTS
None.
ACKNOWLEDGMENTS
We would like to thank the Interprofessional Fund for Agricultural Research and Advisory (FIRCA) for financing the project on which we worked and all the coordinators for their valuable contributions to this work. We would also like to thank the University Péléforo Gon Coulibaly for the close scientific collaboration and also all the anonymous readers of this article.
REFERENCES
Assiri, A. A., Konan, A., N’Guessan, K.F., Kébé, B.I., Kassin, K.E., Couloud, J.Y., Yapo, A. R., Yoro, G.R., and Yao, K.A. (2015). Comparaison de deux techniques de replantation cacaoyère sur antécédents culturaux non-forestiers en Côte d’Ivoire. African Crop Science Journal, 23(4), 365-378. http://dx.doi.org/10.4314/acsj.v23i4.6.
Bayes, T. (1763). An Essay Towards Solving a Problem in the Doctrine of Chances, Philosophical Transactions of The Royal Society of London 53, 370-418. https://doi.org/10.1098/rstl.1763.0053.
Bayes, T. (1958). Studies in the History of Probability and Statistics: IX. Thomas Bayes’s Essay Towards Solving a Problem in the Doctrine of Chances. (Bayes’s Essay in Modernized Notation), Biometrika 45, 296-315. https://doi.org/10.2307/2333180.
Burow, G., Franks, C. D., Xin, Z., Burke, J. J. (2012). Genetic Diversity in a Collection of Chinese Sorghum Landraces Assessed By Microsatellites. American Journal of Plant Sciences 3, 1722-1729. http://dx.doi.org/10.4236/ajps.2012.312210.
Chaabane, R., Babay, E., Sahari, K., Khamassi, K., Ben, S. H., Gharbi, M. S., El, F. M., Deghaies, M., Ben, N. M. (2007). Choix des amorces microsatellites pour l’analyse de la diversité génétique du blé tendre : Acte des 14èmes journées scientifiques sur les résultats de la recherche Agricoles, Communication. http://dx.doi.org/10.13140/2.1.1539.6166.
Clément, D. A., Legnate, H., Seguin, M., Carron, M. P., Leguen, V., Chapuuset, T., and Schultes, R. E. (1990). A Briet Taxonomic View of The Genus Hevea. Malaysian Rubber Research and Development Board Monograph N° 14, Kuala Lumpur.
Compagnon, P. (1986). Le Caoutchouc Naturel : Biologie, Culture, Production. Maisonnneuve Et Larose.
Ghebru, B., Schmidt, R. J., Bennetzen, J. L. (2002). Genetic Diversity of Eritrean Sorghum Landraces Assessed with Simple Sequence Repeat (SSR) Markers. Theoretical and Applied Genetics 105, 229-236. http://dx.doi.org/10.1007/s00122-002-0929-x.
Giovani, S.F.B. (2005). Estimation and Inference in Dynamic Unbalanced Panel-Data Models With A Small Number of Individuals. Stata Journal 5(4), 473-500. https://doi.org/10.1177/1536867X0500500401.
Gnagne, Y.M., Elabo, A.A.E., Okoma, K.K., Konan, E., and Zakra, N. (2014a). Clef d’identification en Jardin à bois de greffes des clones de Hevea brasiliensis vulgarisés en Côte d’Ivoire, Poster.
Keli, J.Z., Obouayeba, S., Zehi, B. (1992). Influence de quelques systèmes vivriers sur le comportement des jeunes hévéas en basse Cote d’Ivoire. Systèmes Agricoles en Afrique 2(1), 41-48.
Kouakou H. M. (2017). Détermination du niveau de fiabilité de la méthode d’identification morphologique en jardin à bois des clones d’Hevea brasiliensis (Euphorbiaceae) diffusés en Côte d’Ivoire par utilisation des marqueurs microsatellites. Mémoire de Master de génétique et amélioration des espèces végétales de l’Université Félix Houphouët Boigny-Abidjan, Côte d’ivoire.
Le Guen, V. F., Doaré, C., Weber, And Seguin, M. (2009). Genetic Structure of Amazonian Populations of Hevea Brasiliensis Is Shaped by Hydrographical Network and Isolation By Distance. Tree Genetics and Genomes 5, 673-683. https://doi.org/10.1007/s11295-009-0218-9.
Obouayeba, S. (2005). Contribution à la détermination de la maturité physiologique de l’écorce pour la mise en saignée d'Hevea brasiliensis Muell. Arg. (euphorbiacée) : normes d'ouverture. Thèse de doctorat Unique de l’Université de Cocody, Côte d’Ivoire.
Peakall, R. and Smouse, P. E. (2012). Genalex 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research an Update. Bioinformatics 28(19), 2537-2539.
Penot, M. E. (2001). Stratégies paysannes et évolution des savoirs : l'hévéaculture agroforestière indonésienne. Thèse de Doctorat de l’Université Montpellier I.
Perrier, X. and Jacquemoud-Collet, J. (2006). DARwin software. Version 6.0, Montpellier.
Pritchard, J.K., Stephens, M. and Donnelly, P., (2000). Inference of Population Structure Using Multilocus Genotype Data. Genetics 155, 945-959. https://doi.org/10.1093/genetics/155.2.945.
Yapi, A. F. (2013). Identification morphologique en Jardin à bois de greffes de clones de Hevea brasiliensis Muell. AGR. (Euphorbiaceae) diffusés en Côte d’Ivoire. Mémoire de master de l’Université de Cocody-Abidjan, Côte d’ivoire.
 This work is licensed under a: Creative Commons Attribution 4.0 International License
This work is licensed under a: Creative Commons Attribution 4.0 International License
© Granthaalayah 2014-2022. All Rights Reserved.